
A Robust Platform to Develop Recombinant Antibodies
For more than a decade, the only way to produce antibodies was by using animal models. But, with new technologies and improved ethical practices, fortunately it is now possible to produce antibodies without animal models. The hyperphage system, which is also known as helper phage technology, was developed by Rondont et al. (Nature Biotechnology 19:75-81, 2001) and is one way to produce recombinant antibodies.
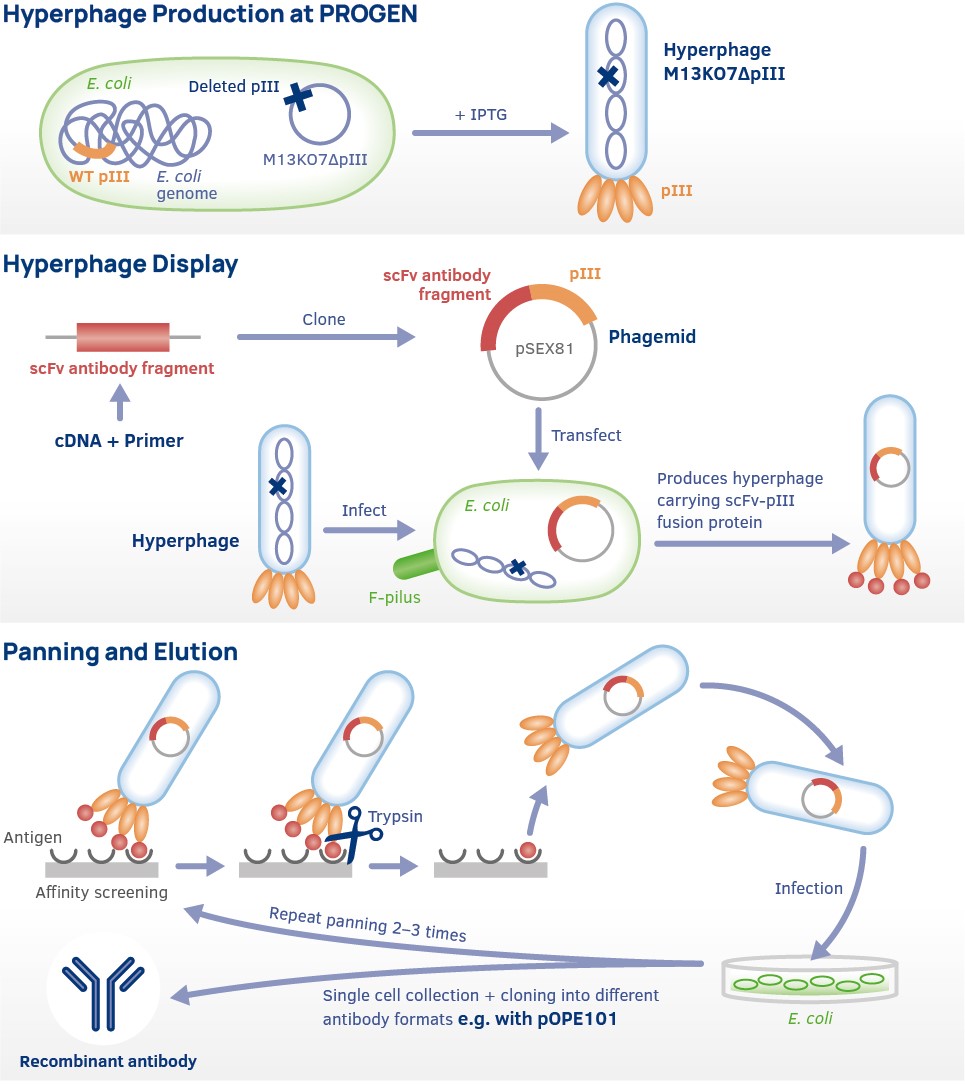
Bacteriophages are viruses that infect bacteria and these are needed for the hyperphage system. They are composed of proteins that encapsulate a DNA or RNA genome and they replicate within a host bacterium after their genome has been injected. The most commonly known bacteriophage for phage display is the M13 phage. The bacteriophage M13 only infects E. coli strains and is part of the group of filamentous phages that refer to the Ff-phages. One of its key features is the possibility to display antibody fragments, proteins or peptides on its minor coat protein, pIII.
The hyperphage system has revolutionized the field of antibody engineering and drug discovery because it improves antibody presentation in phage display by increasing the number of antibodies displayed per phage particle (up to 5 vs. 0.01), compared to normal helper phages. The displayed antibodies on the phages can be used for various applications, such as identifying specific targets or in immune response or disease treatment related research. In cancer research, the hyperphage-packed library could be a tool to discover new tumor markers by panning against cellular surfaces.

Hyperphage Benefits
Exceptional reproducibility: recombinant antibodies are produced in cell cultures or microorganisms, ensuring you get high reproducibility and consistent quality.
Future-proof supply: an animal free platform ensures your recombinant antibodies can be produced time and time again.
Fast production: benefit from a quicker production time in comparison to monoclonal or polyclonal antibody production.
Controlled conditions: ensures precise manipulation, prevents contamination, provides optimal growth, quality control and scalability.
Animal free: recombinant antibodies are produced in bacterial cultures, ensuring you an animal free antibody production guarantee.
Antibody Library Preparation
The first step is to create antibody variable heavy (VH) and light chain (VL) PCR products. Antibody PCR products are ligated into the pSEX81 surface expression vector (phagemid). This creates a fusion protein containing VH and VL combined with the pIII minor capsid protein derived from the M13 bacteriophage.
Suitable Products for this step
- Mouse IgG Library Primer Set
- Human IgG/IgM Library Primer Set
- pSEX81 Surface Expression Phagemid Vector
Hyperphage Display
Hyperphages carry a deletion in the pIII gene. They are generated by an E. coli packaging cell line producing functional pIII. The resulting hyperphages carry functional pIII on their surface but lack the pIII gene in their genome. These hyperphages can be used to infect bacteria with a phagemid library. Due to the resulting display phages carrying several copies of the antibody or peptide on its surface, panning efficiency is increased dramatically.
Suitable Products for this step
Quantification
The anti-M13/fd/F1 specifically detects the phage outer surface protein pVIII. Using the Phage Titration ELISA, a standard curve is prepared using a dilution series of phage suspension of known titer. The particle number is estimated in comparision to the standard curve.
Suitable Products for this step
Expression of functional recombinant single-chain Fv antibody fragments in E. coli
The identified clones of interest can easily be produced by E. Coli. This is done by transforming the bacteria with the pOPE101 Expression Vector, which includes the desired antibody fragment. As the expression vector contains a c-Myc and His-tag, the expressed antibody fragment can be purified by metal chelation or affinity purification.
Suitable Products for this step
References
You can find protocols and original papers on the successful use of the hyperphage system here.


